LeXPrep EZ RNA Library Preparation Module
- #LX02441
- #LX02442
LeXPrep EZ RNA Library Preparation Kit is a high-speed solution engineered for mainstream NGS platforms. It efficiently converts 2.5–500 ng of RNA—sourced from cells, fresh tissues, blood, microbes, or FFPE—into premium sequencing libraries. Whether you are performing gene expression profiling, fusion gene analysis, or RNA pathogen detection, LeXPrep delivers the reliability your research demands.
- Product Details
- Kit Content
- FAQ
- Ordering Information
- Resources
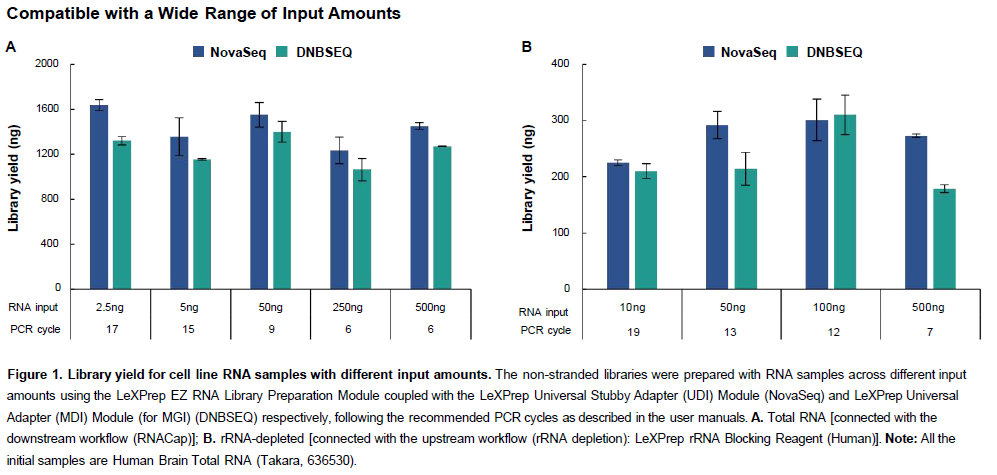
Broad Versatility
- Flexible Input: Supports 2.5–500 ng of initial RNA.
- Sample Agnostic: Compatible with diverse sample types and qualities.
- Customizable: Supports both stranded and non-stranded library prep.
- Universal Compatibility: Adapters work with all mainstream NGS platforms.
- Modular: Seamlessly integrates with existing upstream/downstream workflows.
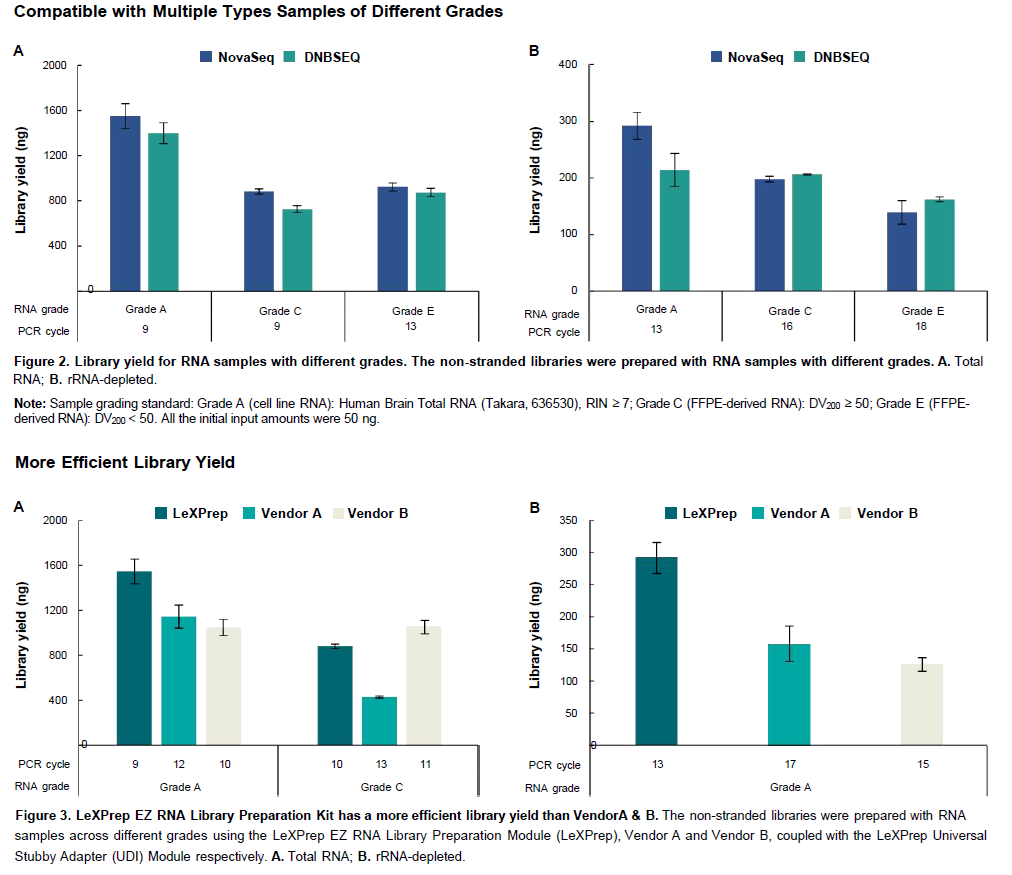
Superior Performance
- High Efficiency: Higher library yields and excellent complexity.
- Precision: More uniform transcript coverage and high strand specificity.
- Deep Detection: Superior sensitivity for low-abundance transcripts and fusion genes.
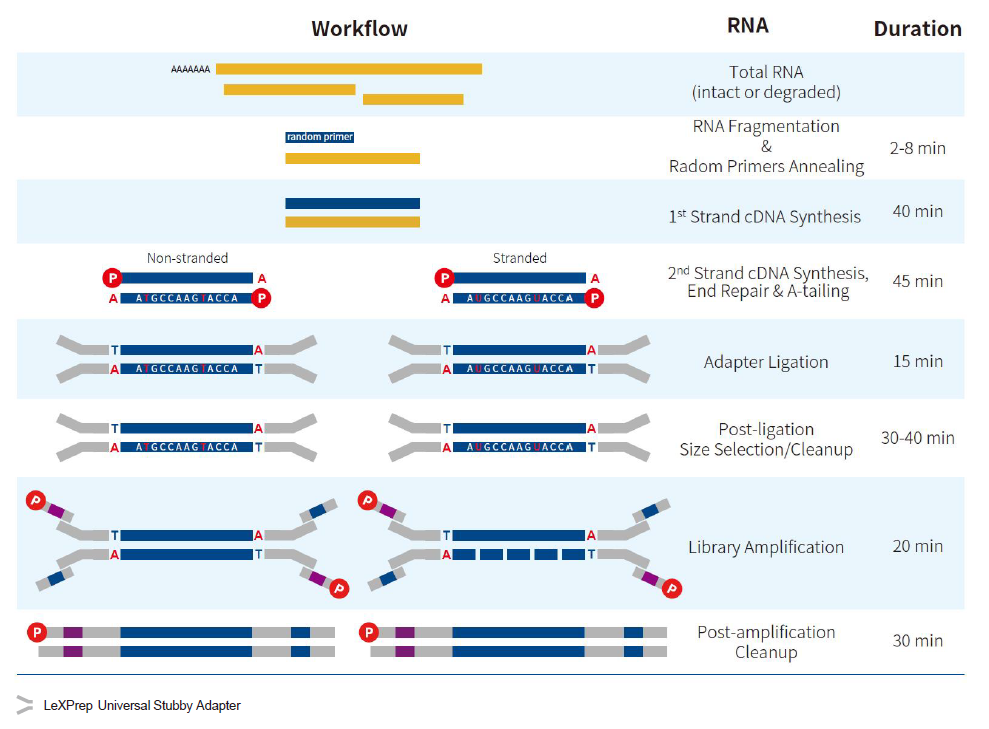
Rapid Workflow
- Efficiency: Combines 2nd-strand synthesis, End Repair, and A-tailing into a single step.
- Fast & Simple: Streamlined, user-friendly protocol significantly reduces hands-on time and total duration.
| Package | Color Of Tube Cap | Component | Volume (24 rxn) | Volume (96 rxn) |
| Box 1 | ● | Random Primer II | 30 µL | 115 µL |
| ● | 1st Strand Buffer | 120 µL | 460 µL | |
| ● | 1st Strand Buffer (stranded) | 120 µL | 460 µL | |
| ● | 1st Strand Enzyme | 60 µL | 230 µL | |
| ● | 2nd Strand & EA Buffer | 750 µL | 2×1500 µL | |
| ● | 2nd Strand & EA Buffer (with dUTP) | 750 µL | 2×1500 µL | |
| ● | 2nd Strand & EA Enzyme | 120 µL | 460 µL | |
| ● | Ligation Buffer | 750 µL | 2×1500 µL | |
| ● | DNA Ligase | 65 µL | 250 µL | |
| ● | 2X HiFi PCR Master Pro Mix | 720 µL | 2×1600 µL | |
| ● | TE Solution | 1 mL | 4 mL | |
| / | Nuclease Free Water | 2.5 mL | 8 mL | |
| Box 2 | / | LeXPrep SP Beads | 4 mL | 15 mL |
Low library yields are typically caused by three primary factors:
- RNase Contamination: RNA is highly susceptible to degradation. Solution: Maintain a strictly nuclease-free environment. Use RNase-free certified consumables and perform all steps in a dedicated clean space to prevent degradation that inhibits reverse transcription.
- Sample Impurity: Residual contaminants (e.g., phenol, ethanol, guanidine salts, or chelators) can inhibit enzymatic activity. Solution: We recommend an additional RNA purification step using LeXPrep SP Beads (2X ratio) and eluting in Nuclease-Free Water to ensure optimal enzyme performance.
- Low Initial Quality: Severely degraded samples naturally produce lower yields. Solution: For low-quality or highly fragmented samples, increase the number of PCR enrichment cycles as recommended in the technical manual.
Yes. The LeXPrep EZ RNA Library Preparation Kit is engineered for high versatility and is compatible with various sample types, including cells, fresh tissues, and FFPE-derived RNA.
For highly fragmented FFPE samples (e.g., DV200 < 20), reverse transcription efficiency may be reduced. In these cases, we recommend:
- Increasing the number of PCR cycles.
- Increasing the initial RNA input amount—especially for targeted capture applications—to maximize complexity and capture efficiency.
The LeXPrep EZ RNA Library Preparation Kit is widely applicable to various starting materials, including:
- Total RNA (from multiple species and grades)
- Poly(A)-enriched mRNA
- rRNA-depleted RNA
Note on Input: Because mRNA content varies significantly between different tissue types and sources, ensure that your total RNA input is sufficient to yield the required amount of mRNA for high-complexity library construction.
The primary difference lies in the preservation of the transcriptional directional information:
- Stranded (Strand-Specific) Libraries: These retain the original orientation of the transcript. This allows researchers to determine whether a read originated from the sense or antisense strand. Stranded RNA-Seq is essential for accurate gene quantification, identifying antisense transcripts, and resolving overlapping gene structures.
- Non-Stranded (General) Libraries: Directional information is lost during second-strand cDNA synthesis. While suitable for basic expression profiling, non-stranded libraries cannot distinguish between sense and antisense transcription.
Recommendation: For comprehensive studies of gene regulation and structure, stranded library preparation is the preferred gold standard for higher-resolution data.
| Product | Catalog |
|---|---|
| LeXPrep EZ RNA Library Preparation Module, 24 rxn | LX02441 |
| LeXPrep EZ RNA Library Preparation Module, 96 rxn | LX02442 |
For inquiries or to request a formal quotation, please contact us at support@lexigenbio.com
