LeXOncofusion (for RNA) Panel
- #LX01512
- #LX01511
- #LX01513
LeXOncoFusion (RNA) Panel: Definitive Fusion Discovery & Transcriptomic Insight
The LeXOncoFu (RNA) Panel is a high-performance targeted enrichment solution engineered for the precise identification and validation of gene fusions in solid tumor research. By operating at the transcript level, this panel provides functional evidence of genomic rearrangements, bridging the gap between DNA-level structural variants and their actual biological impact.
- Product Details
- Kit Content
- FAQ
- Ordering Information
- Resources
Targeted Precision for Solid Tumors
The panel focuses on 105 high-impact genes curated for their high clinical relevance and frequent involvement in oncogenic fusions. To ensure no critical data is missed, the probe design offers exhaustive coverage across:
- Transcript Coding Regions (CDS): Complete enrichment of all coding sequences as defined by RefSeq 109.
- Strategic UTR Coverage: Targeted inclusion of critical Untranslated Regions (UTRs) where fusion breakpoints and regulatory disruptions frequently occur.
Triple-Layer Data Insights in a Single Assay
The LeXOncoFu system enables researchers to extract multidimensional information from a single RNA sample, maximizing data yield from often-limited clinical specimens:
- Fusion Detection: Definitively identify known and novel fusion events with their specific partner genes and isoforms.
- Expressed Mutation Analysis: Detect single nucleotide variants (SNVs) and indels within active transcripts to assess the functional mutation load.
- Gene Expression Profiling: Quantify transcript abundance to identify signature patterns of oncogenic transformation and pathway activation.
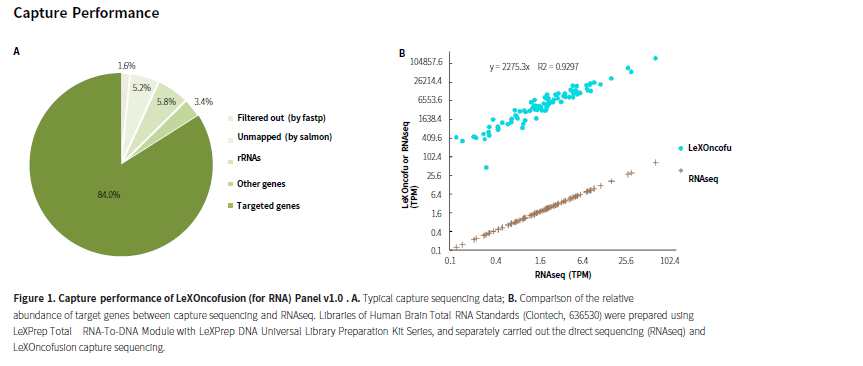
| ABL1*† | ABL2 | AKT3 | ALK | ARHGAP26 | AXL | BCL2† | BCOR* | BRAF | BRD3* | BRD4* | CAMTA1 |
| CCNB3* | CCND1 | CIC† | COL6A3 | CSF1R | DNAJB1 | EGFR | EML4 | EPC1* | ERBB2† | ERG* | ESR1 |
| ETS1 | ETV1* | ETV4 | ETV5* | ETV6* | EWSR1 | FGFR1 | FGFR2 | FGFR3 | FGR* | FLI1 | FLT3 |
| FOSB | FOXO1 | FUS | GLI1 | HMGA2 | INSR | JAK2* | JAK3 | JAZF1 | KIF5B† | KIT† | KMT2A |
| MAML2 | MAST1 | MAST2 | MEAF6 | MET* | MLLT3 | MSH2 | MSMB† | MUSK | MYB | MYC | NCOA1* |
| NCOA2* | NOTCH1 | NOTCH2 | NOTCH3† | NR4A3* | NRG1 | NTRK1 | NTRK2 | NTRK3 | NUMBL | NUTM1 | PAX3 |
| PAX7 | PDGFB | PDGFRA | PDGFRB† | PHF1* | PIK3CA* | PKN1 | PLAG1* | PPARG* | PRKACA | PRKCA | PRKCB |
| RAF1 | RARA* | RELA | RET | ROS1 | RSPO2* | RSPO3 | SS18 | STAT6* | TAF15 | TCF12† | TCF3 |
| TERT | TFE3 | TFEB | TFG | THADA | TMPRSS2*† | TP53* | USP6* | YWHAE* |
*Covers all 5′-UTRs; †Covers all 3′-UTRs。
| Package | Color Of Tube Cap | Component | Volume | Package/Storage |
| LX01512 | ● | LeXOncofusion (for RNA) Panel v1.0, 16 rxn | 70 μL | -20℃ |
| LX01511 | ● | LeXOncofusion (for RNA) Panel v1.0, 96 rxn | 415 μL | -20℃ |
While DNA sequencing identifies potential rearrangements, only RNA sequencing can confirm if those rearrangements lead to functional, aberrant transcripts. The LeXOncoFusion (RNA) Panel is the essential tool for researchers seeking to validate driver fusions with high sensitivity, even in samples with low tumor purity or complex genomic backgrounds.
Because our probes cover the entire coding sequence and critical UTRs of the 105 target genes, the panel can capture fusions involving both known and novel partner genes. This “anchor-based” approach ensures that if a target gene is rearranged, its partner will be enriched and identified during bioinformatic analysis.
Yes. The LeXOncoFusion workflow is optimized for the challenges of solid tumor research, including the use of RNA extracted from Formalin-Fixed Paraffin-Embedded (FFPE) tissues. The targeted capture approach significantly improves the signal-to-noise ratio compared to whole-transcriptome methods, providing reliable results from fragmented samples.
| Product | Catalog |
|---|---|
| LeXOncoFusion (for RNA) Panel v1.0, 16 rxn | LX01512 |
| LeXOncoFusion (for RNA) Panel v1.0, 96 rxn | LX01511 |
| LeXOncoFusion (for RNA) Panel v1.0, 480 rxn | LX01513 |
For inquiries or to request a formal quotation, please contact us at support@lexigenbio.com
